Specify images/parameters in Seg2D+Link
Parameter panels
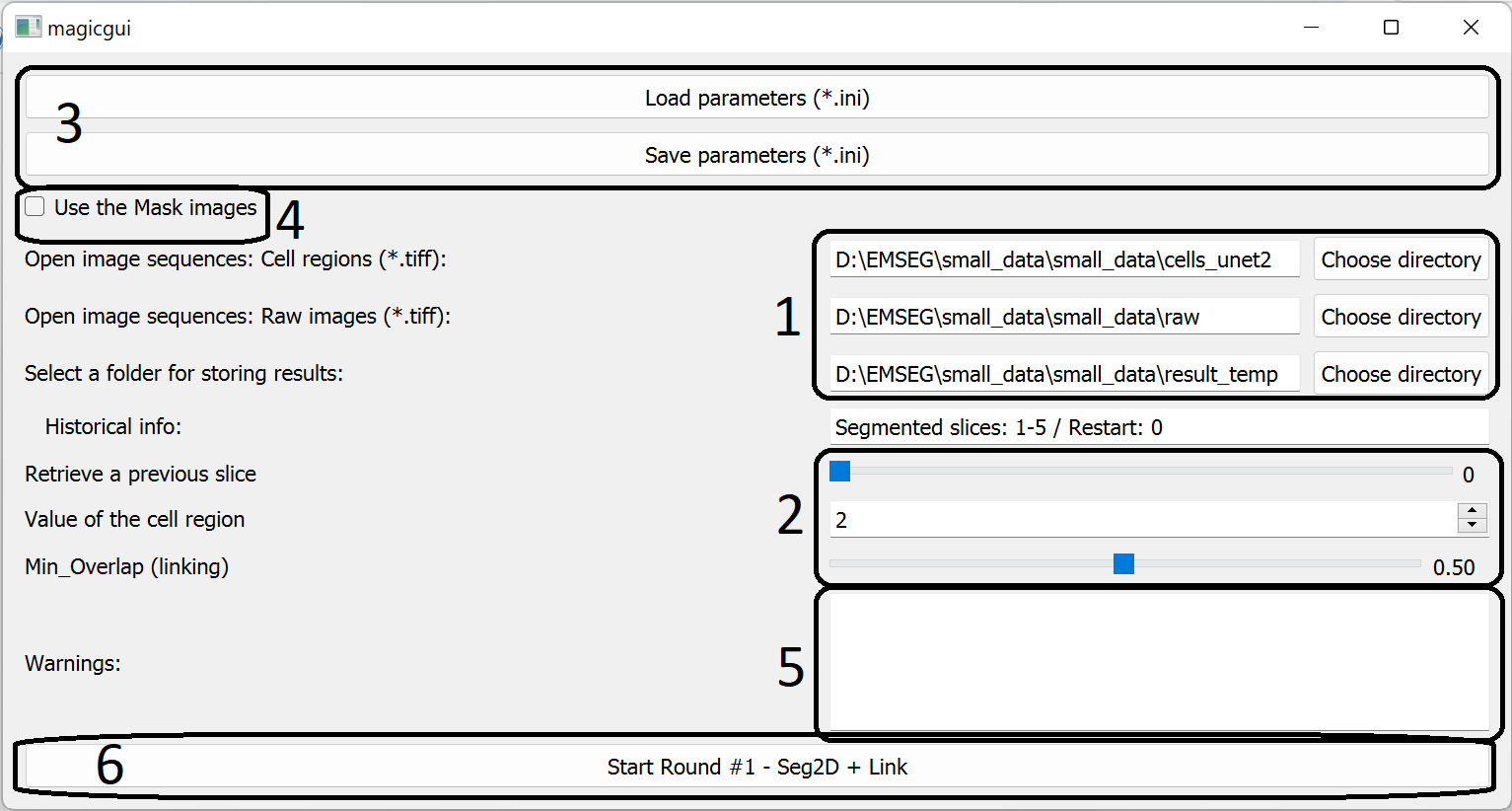
1. Paths of images/results
- Users should specify three folders:
- containing the cell/non-cell prediction images (2D Tiff images);
- containing the raw images (2D Tiff images);
- to store the segmentation results.
2. Parameters
- Users should specify following parameters:
- Slice number to be retrieved.
- Default value is the last slice that have been segmented.
- The segmentation results after the specified slice number will be deleted.
- If set it to 0, the program will restart the segmentation and remove all previous segmentation results.
- Value of the cell region.
- Set it according to the cell/non-cell images.
- The minimal overlap coefficient.
- This is a custom threshold used for linking cells across slices.
- Supposing there are two cells X and Y in two adjacent slices, their overlap coefficient is: $$overlap(X,Y)=\frac{Area(X\cap Y)}{min(Area(X),Area(Y))} $$
- If \(overlap(X, Y) > threshold\), cells X and Y will be linked.
- Slice number to be retrieved.
3. Save/Load the paths/parameters
- Users can save the specified parameters in a .ini file and reload it later to avoid having to set the paths/parameters every time the software is launched.
- Users can modify other advanced parameters (e.g. the maximal number of steps of cached actions) in the saved .ini files (Explanations).
4. Use Mask images
This option allows users to segment cells in regions of interest (ROI) defined by users.
When it is selected, the GUI will change as follows:
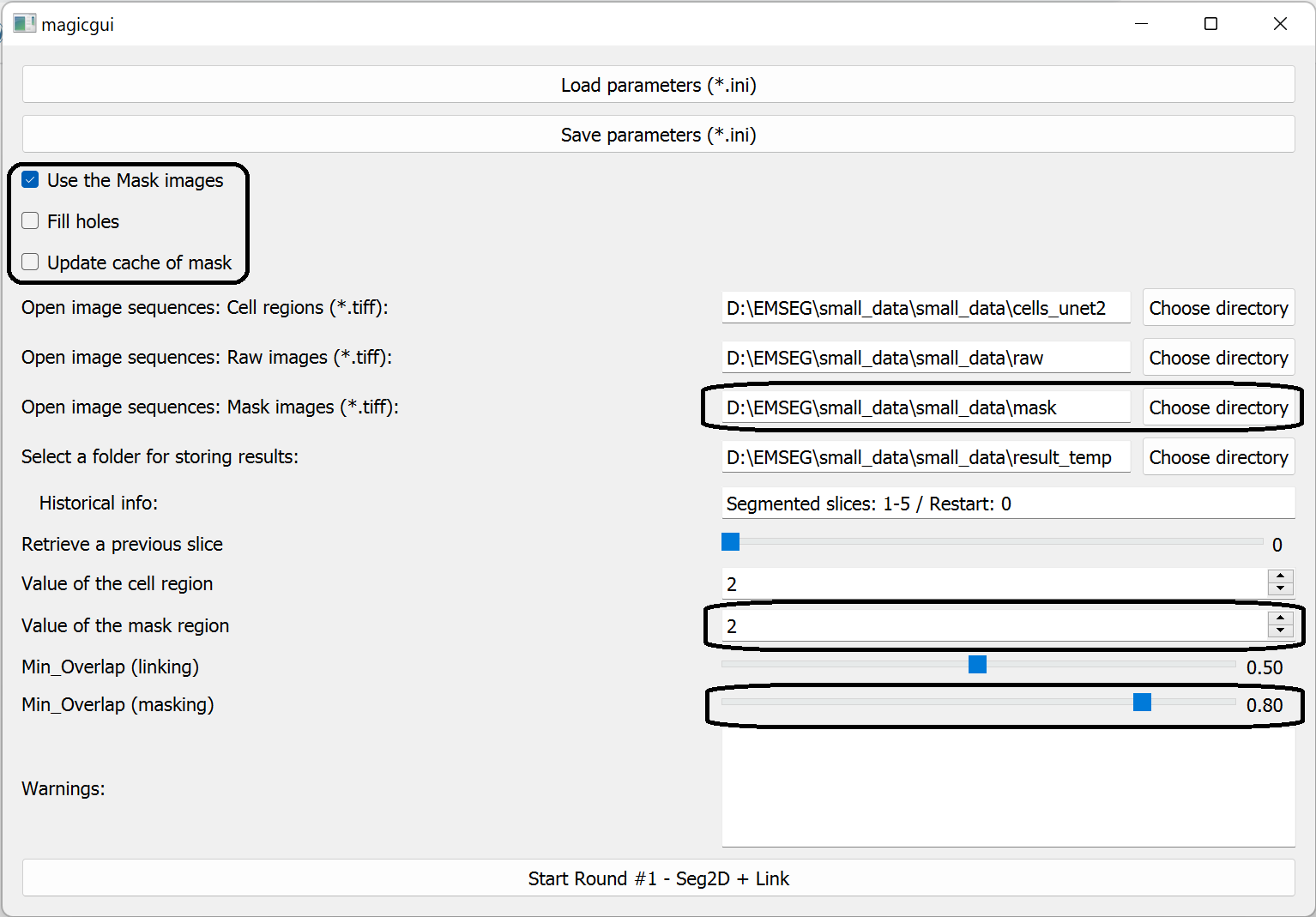
Users should further specify following parameters:
- Fill holes.
- Force the program to automatically fill the holes in the user-defined ROI. The calculation will take a long time when processing large image (but only once).
- Update cache of mask.
- The cache is a .npy file of the original/holes-filled mask image. It was created to avoid repeated calculations after launching the software.
- Check this option will force the program to re-make the cache file, which will take a long time.
- If it is not checked, the program will try to load the mask data from the existed cache file.
- Path for mask images.
- Specify the folder containing the mask images (2D Tiff images);
- Value of the mask region.
- Set this to the value of the user-defined ROI in mask images.
- The minimal overlap.
- This is a custom threshold for ignoring cells outside of the ROI..
- Assuming a cell A was partially located within the ROI, then the overlap of A within ROI is: $$overlap(A)=\frac{Area(X\cap ROI)}{Area(A)} $$
- If \(overlap(A) < threshold\), Cell A will be removed from the segmentation result.
5. Warning information
If the images or specified folders are not found, the warning information will be displayed here.
6. Start the Seg2D+Link Module
Press this button to launch the Seg2D+Link module.